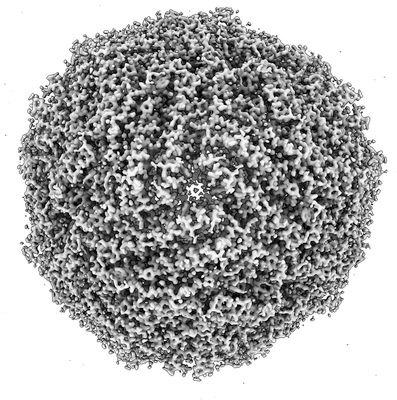
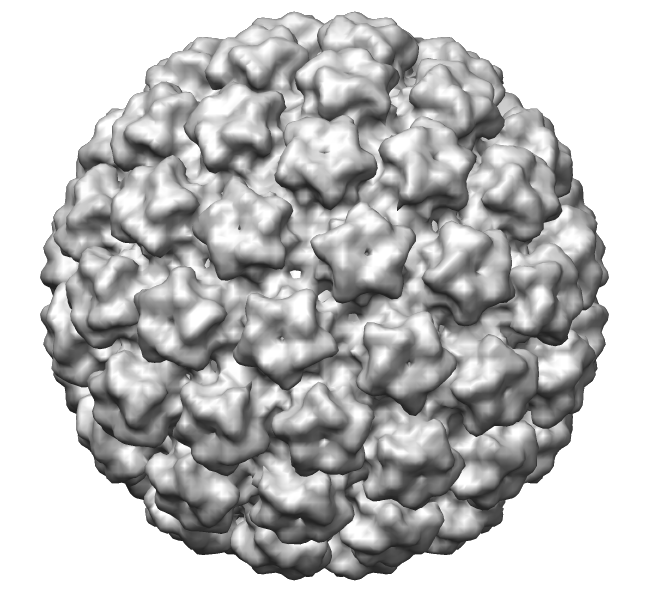
Mouse apoferritin image processing in Scipion 3
This tutorial presents a complete workflow of Cryo-EM single particles inside Scipion 3. It demonstrates the combination of different EM-packages with high focus on Xmipp highres.
Guide:
apoferritin image processing tutorial in scipion3
You may download a fully solved compressed project (Example_10248_Scipion3.tgz) of this tutorial from here.
Workflow: download
Relion in Scipion
This tutorial is focused on using Relion 5 single particle workflow inside Scipion. It follows the processing pipeline described for Relion 5.0 tutorial with Beta-galactosidase data.
Guide: scipion_tutorial_relion.pdf
Full SPA processing video tutorial
4 video tutorials in a list to go from movies to a 3D volume using betagalactosidase data.
Part1: Micrograph processing
Part2: Particle picking
Part3: 2D classification and initial volume
Part4: 3D Reconstruction and validation
Take a look at our tutorial videos in the Scipion youtube channel.
Know more about the theory and practice behind Image Processing in EM.
Initial model estimation
This tutorial uses different methods to obtain an initial 3D map. Methods covered are RCT, Ransac, Eman and Reconstruct Significant.
Initial volume validation by SAXS
This tutorial provides a guide to the validation of the initial volume estimations using SAXS curves in Scipion.
Guide: scipion_tutorial_SAXS.pdf
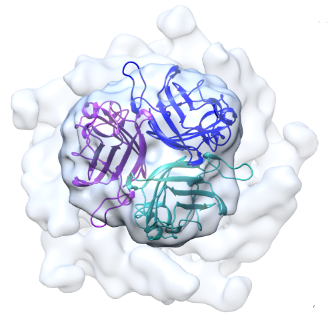
Localized reconstruction in Scipion
In this tutorial, we provide a step-by-step guide for handling symmetry mismatches in single-particle analysis using a new plugin called LocalRec (Ilca et al. 2015) in Scipion (Abrishami et al. 2020).